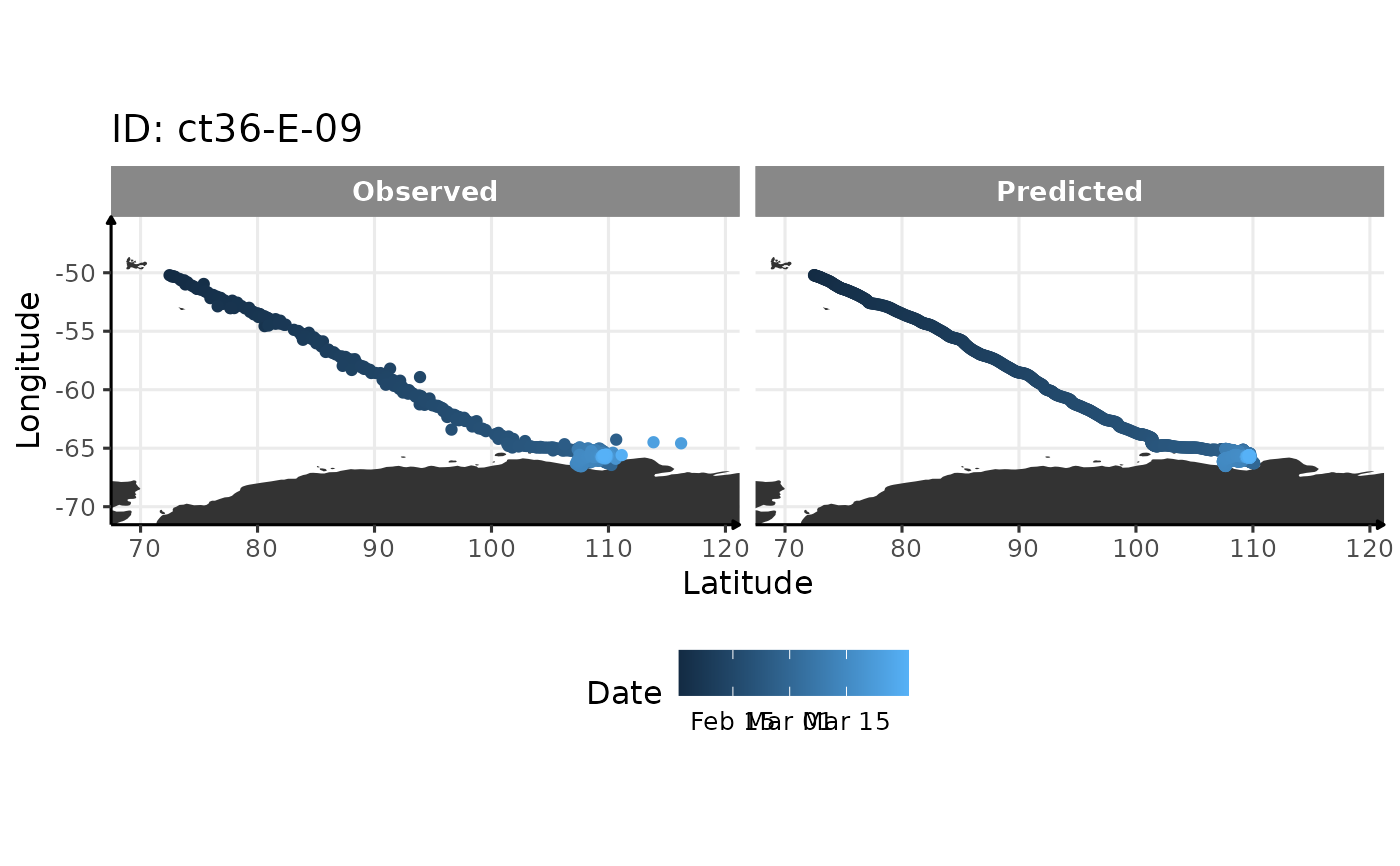
Treat location data with a continuous-time state-space model
location_treatment.RdUsing fit_ssm function from foieGras package, this function "clean" the
location data to be used for further analysis at the dive scale.
Usage
location_treatment(
data,
model = "crw",
time.step = 1,
vmax = 3,
with_plot = FALSE,
export = NULL
)Arguments
- data
Dataset of observation, usually the file \*Argos.csv or \*Location.csv files
- model
Choose to fit either a simple random walk (
rw) or correlated random walk (crw) as a continuous-time process model- time.step
options: 1) the regular time interval, in hours, to predict to; 2) a vector of prediction times, possibly not regular, must be specified as a data.frame with id and POSIXt dates; 3) NA - turns off prediction and locations are only estimated at observation times.
- vmax
The max travel rate (m/s) passed to sda to identify outlier locations
- with_plot
A diagnostic plot
- export
To export the new generated dataset
Examples
# load library
library(foieGras)
library(data.table)
# run this function on sese1 dataset included in foieGras package
output <- location_treatment(copy(sese1), with_plot = TRUE)
#> fitting crw...
#>
pars: 0 0 0 0
pars: -0.07009 -0.67701 -0.73263 -0.00241
pars: 0.80735 -0.49698 -1.10459 0.24116
pars: 0.79068 -0.93408 -0.86323 0.26125
pars: 1.17074 -0.84805 -0.95396 0.56112
pars: 0.94797 -0.82123 -0.95447 0.38458
pars: 0.71902 -0.86148 -1.03497 0.40682
pars: 0.90772 -0.86547 -0.99023 0.38642
pars: 0.86034 -0.81931 -0.96958 0.37881
pars: 0.84471 -0.83656 -0.93133 0.40567
pars: 0.86034 -0.81931 -0.96958 0.37881